Contents
PRISM Algorithm 2
%matplotlib widget
import py4DSTEM
import abtem
import ase
import ipywidgets
import numpy as np
import matplotlib.pyplot as plt
from matplotlib.gridspec import GridSpec
abtem.config.set({"dask.lazy":False});Multislice¶
Si3N4_crystal = ase.Atoms(
"Si6N8",
scaled_positions=[
(0.82495, 0.59387, 0.75),
(0.23108, 0.82495, 0.25),
(0.59387, 0.76892, 0.25),
(0.40614, 0.23108, 0.75),
(0.76892, 0.17505, 0.75),
(0.17505, 0.40613, 0.25),
(0.66667, 0.33334, 0.75),
(0.33334, 0.66667, 0.25),
(0.66986, 0.70066, 0.75),
(0.96920, 0.66986, 0.25),
(0.70066, 0.03081, 0.25),
(0.29934, 0.96919, 0.75),
(0.33015, 0.29934, 0.25),
(0.03081, 0.33014, 0.75),
],
cell=[7.6045, 7.6045, 2.9052, 90, 90, 120],
pbc=True
)
Si3N4_orthorhombic = abtem.orthogonalize_cell(Si3N4_crystal)
Si3N4_orthorhombic *= (3,2,17)potential = abtem.Potential(
Si3N4_orthorhombic,
# sampling = 0.2,
gpts = (128,144),
slice_thickness = Si3N4_orthorhombic.cell[2,2]/32,
parametrization = 'lobato',
projection = 'finite',
).build(
)s_matrix_1 = abtem.SMatrix(
potential=potential,
energy=300e3,
semiangle_cutoff=20,
downsample=None,
interpolation=1,
).build(
)
delta = 26
arrays = s_matrix_1.waves[:5*delta:delta].array
wavevectors = s_matrix_1.waves[:5*delta:delta].ensemble_axes_metadata[0].values
titles = [f"q=({kx:.3f},{ky:.3f})"+r" $\AA^{-1}$" for kx,ky in wavevectors]
with plt.ioff():
py4DSTEM.show_complex(
arrays,
title=titles,
vmin=0,vmax=1,
ticks=False,
cbar=False,
axsize=(3,3)
)
plt.gcf() 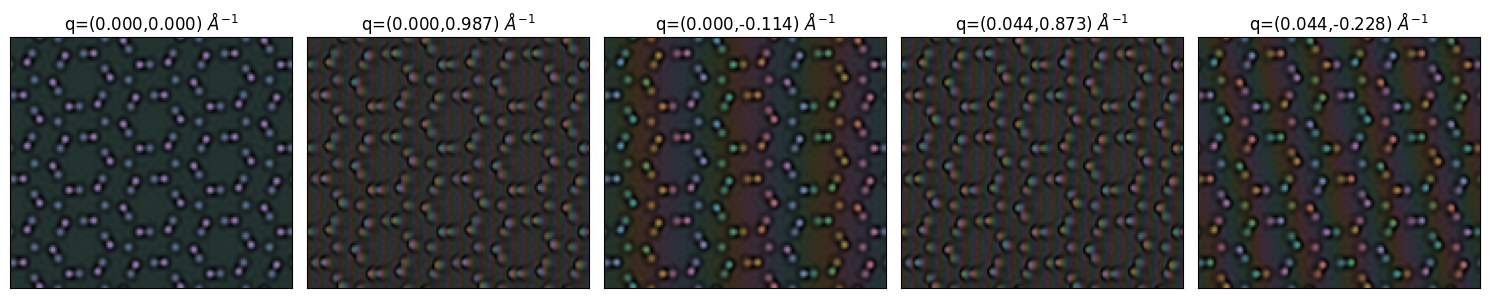
Interpolation Factor¶
s_matrix_2 = abtem.SMatrix(
potential=potential,
energy=300e3,
semiangle_cutoff=20,
downsample=None,
interpolation=2,
).build(
)
s_matrix_3 = abtem.SMatrix(
potential=potential,
energy=300e3,
semiangle_cutoff=20,
downsample=None,
interpolation=3,
).build(
)
s_matrix_4 = abtem.SMatrix(
potential=potential,
energy=300e3,
semiangle_cutoff=20,
downsample=None,
interpolation=4,
).build(
)
s_matrix_6 = abtem.SMatrix(
potential=potential,
energy=300e3,
semiangle_cutoff=20,
downsample=None,
interpolation=6,
).build(
)The interpolation factor does not exactly divide 'gpts', normalization may not be exactly preserved.
The interpolation factor does not exactly divide 'gpts', normalization may not be exactly preserved.
The interpolation factor does not exactly divide 'gpts', normalization may not be exactly preserved.
The interpolation factor does not exactly divide 'gpts', normalization may not be exactly preserved.
probe_arrays = [
s.reduce(
scan=abtem.CustomScan(np.array(potential.extent)/2),
ctf=abtem.CTF(defocus=100),
detectors=abtem.WavesDetector()
).array[0]
for s in (s_matrix_1,s_matrix_2,s_matrix_3,s_matrix_4,s_matrix_6)
]
probe_fourier_arrays = [np.fft.fftshift(np.fft.fft2(np.fft.ifftshift(arr))) for arr in probe_arrays]
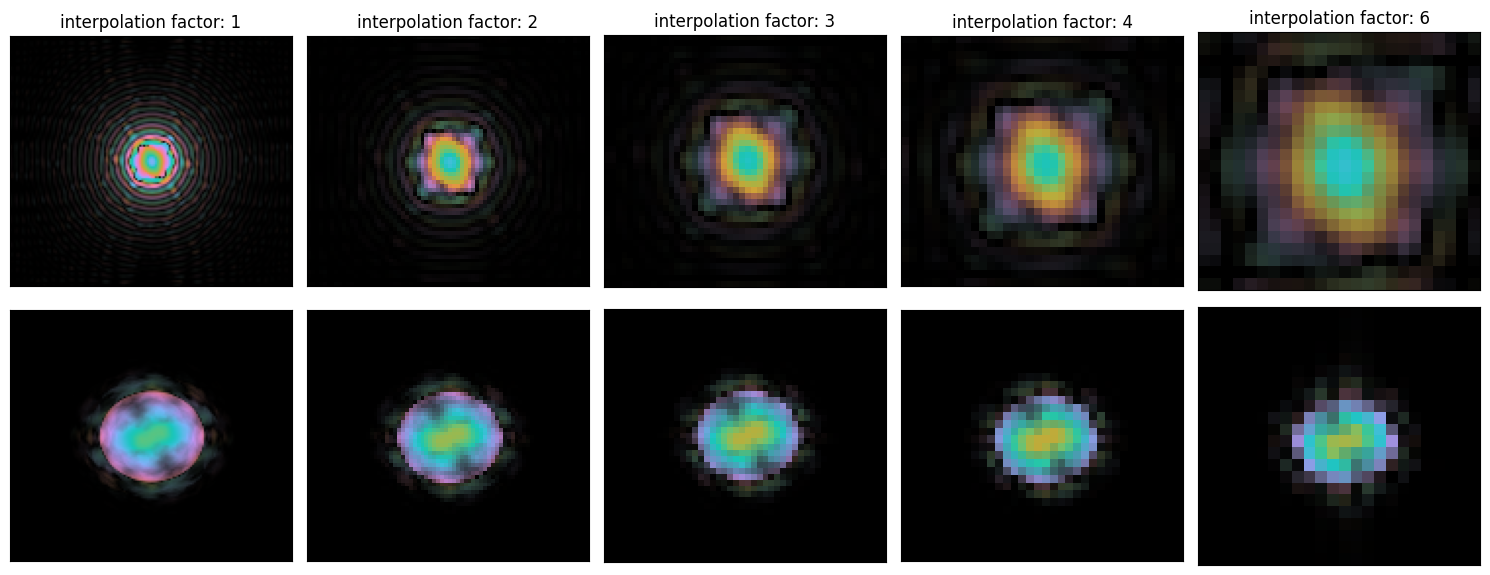
probe_titles = [f"interpolation factor: {f}" for f in (1,2,3,4,6)]with plt.ioff():
py4DSTEM.show_complex(
[
probe_arrays,
probe_fourier_arrays
],
ticks=False,
cbar=False,
title=probe_titles,
axsize=(3,3),
)
plt.gcf()