Contents
Electron Wavefunctions
%matplotlib widget
import numpy as np
import matplotlib.pyplot as plt
from mpl_toolkits.axes_grid1 import make_axes_locatable, ImageGrid
from IPython.display import display, clear_output
import ipywidgets
import abtem
import py4DSTEM
abtem.config.set({"dask.lazy":False});PSF/CTF¶
symbols = [symbol for symbol in abtem.transfer.polar_symbols if symbol[0] == "C"]
nm = [(int(c[-2]), int(c[-1])) for c in symbols]
dicts = [{symbol: 50**n} for symbol, (n,m) in zip(symbols,nm)]
ctf_objects = [abtem.CTF(semiangle_cutoff=30,energy=100e3,**coefs) for coefs in dicts]
ctfs = [ctf.to_diffraction_patterns(max_angle=30).array for ctf in ctf_objects]
psfs = [ctf.to_point_spread_functions(gpts=256,extent=40).crop(extent=(10,10),offset=(15,15)).array for ctf in ctf_objects]def plot_coefficients(nm, arrays):
nrows, ncols = (5, 7)
dpi = 72
with plt.ioff():
# fig = plt.figure(figsize=(675/dpi,562.5/dpi),dpi=dpi)
fig = plt.figure(figsize=(720/dpi,600/dpi),dpi=dpi)
axes = ImageGrid(
fig,
111,
nrows_ncols=(nrows, ncols),
**{"axes_pad": 0.0},
)
axes = np.array(axes).reshape((5, 7))
for array, (n, m) in zip(arrays, nm):
ax = axes[(n - 1, m)]
rgb = py4DSTEM.visualize.Complex2RGB(array,vmin=0,vmax=1)
ax.imshow(rgb)
for ax in axes.ravel():
ax.set_xticks([])
ax.set_yticks([])
ax.set_xlabel("")
ax.set_ylabel("")
ax.spines["top"].set_visible(False)
ax.spines["right"].set_visible(False)
ax.spines["bottom"].set_visible(False)
ax.spines["left"].set_visible(False)
for i, ax in enumerate(axes[:, 0]):
ax.set_ylabel(f"$n={i + 1}$", fontsize=12)
for i, ax in enumerate(axes[0, :]):
ax.set_title(f"$m={i}$", fontsize=12)
return figctfs_fig = plot_coefficients(nm,ctfs)
# ctfs_fig.savefig("../figures/CTFs_table.png",bbox_inches="tight",transparent=True)
ctfs_fig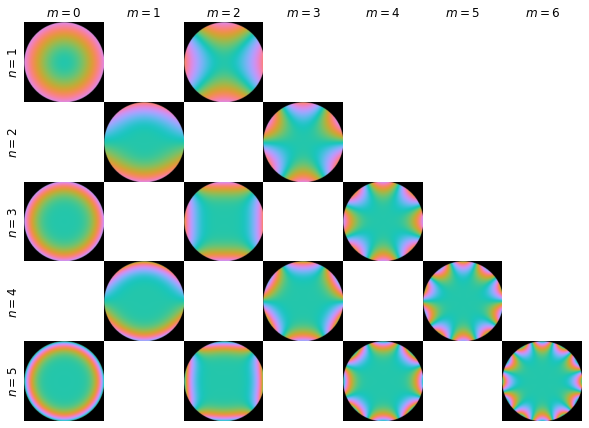
psfs_fig = plot_coefficients(nm,psfs)
# psfs_fig.savefig("../figures/PSFs_table.png",bbox_inches="tight",transparent=True)
psfs_fig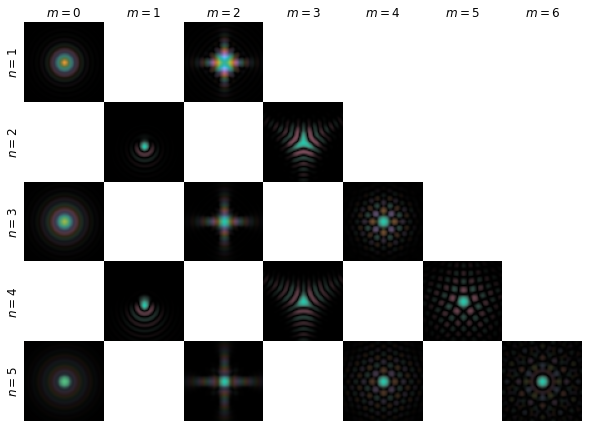
Apertures¶
def return_probe_arrays(semiangle_cutoff,aberrations=None,**kwargs):
""" """
ctf = abtem.CTF(
semiangle_cutoff=semiangle_cutoff,
sampling=(0.125,0.125),
gpts=(128,128),
energy=300e3,
aberration_coefficients=aberrations,
**kwargs,
)
fourier_probe = ctf.to_diffraction_patterns(
gpts=ctf.gpts
).array
complex_probe = ctf.to_point_spread_functions(
gpts=ctf.gpts,
extent=ctf.extent
).array
return ctf, fourier_probe, complex_probe
ctf, fourier_probe, complex_probe = return_probe_arrays(
semiangle_cutoff=20,
)
dpi = 72
with plt.ioff():
fig,axs = plt.subplots(1,2,figsize=(675/dpi,325/dpi),dpi=dpi)
bar_common = {"Nx":128,"Ny":128,"labelsize":8}
rgb_fourier_probe = py4DSTEM.visualize.Complex2RGB(
fourier_probe,
vmin=0,vmax=1,
)
im_fourier = axs[0].imshow(rgb_fourier_probe)
bar_fourier= bar_common | {"pixelsize":ctf.angular_sampling[0],"pixelunits":"mrad"}
rgb_complex_probe = py4DSTEM.visualize.Complex2RGB(
complex_probe,
vmin=0,vmax=1,
)
im_complex = axs[1].imshow(rgb_complex_probe)
bar_complex= bar_common | {"pixelsize":ctf.sampling[0],"pixelunits":r"$\AA$"}
bars = [bar_fourier, bar_complex]
titles= ["Fourier-space probe", "real-space probe"]
for ax, bar, title in zip(axs, bars, titles):
ax.set(xticks=[],yticks=[],title=title)
divider = make_axes_locatable(ax)
ax_cb = divider.append_axes("right", size="5%", pad="2.5%")
py4DSTEM.visualize.add_colorbar_arg(ax_cb)
py4DSTEM.visualize.add_scalebar(ax,bar)
fig.tight_layout()
fig.canvas.resizable = False
fig.canvas.header_visible = False
fig.canvas.footer_visible = False
fig.canvas.toolbar_visible = True
fig.canvas.layout.width = '675px'
fig.canvas.layout.height = '350px'
fig.canvas.toolbar_position = 'bottom'def update_aperture(change):
""" """
semiangle_cutoff = change['new']
ctf, fourier_probe, complex_probe = return_probe_arrays(
semiangle_cutoff=semiangle_cutoff,
)
rgb_fourier_probe = py4DSTEM.visualize.Complex2RGB(
fourier_probe,
vmin=0,vmax=1,
)
im_fourier.set_data(rgb_fourier_probe)
rgb_complex_probe = py4DSTEM.visualize.Complex2RGB(
complex_probe,
vmin=0,vmax=1,
)
im_complex.set_data(rgb_complex_probe)
fig.canvas.draw_idle()
return None
style = {
'description_width': 'initial',
}
layout = ipywidgets.Layout(width='450px',height='30px')
semiangle_slider = ipywidgets.FloatSlider(
min=5,
max=50,
step=0.5,
value=20,
layout=layout,
style=style,
description="convergence semi-angle [mrad]",
)
semiangle_slider.observe(update_aperture, 'value')display(
ipywidgets.VBox(
[
semiangle_slider,
fig.canvas
],
layout=ipywidgets.Layout(
align_items="center"
)
)
)VBox(children=(FloatSlider(value=20.0, description='convergence semi-angle [mrad]', layout=Layout(height='30px…Aberration Function¶
ab_ctf, ab_fourier_probe, ab_complex_probe = return_probe_arrays(
semiangle_cutoff=20,
defocus=100,
astigmatism=50,
astigmatism_angle=np.pi/4,
)
dpi = 72
with plt.ioff():
ab_fig,ab_axs = plt.subplots(1,2,figsize=(675/dpi,325/dpi),dpi=dpi)
ab_rgb_fourier_probe = py4DSTEM.visualize.Complex2RGB(
ab_fourier_probe,
vmin=0,vmax=1,
)
ab_im_fourier = ab_axs[0].imshow(ab_rgb_fourier_probe)
ab_rgb_complex_probe = py4DSTEM.visualize.Complex2RGB(
ab_complex_probe,
vmin=0,vmax=1,
)
ab_im_complex = ab_axs[1].imshow(ab_rgb_complex_probe)
for ax, bar, title in zip(ab_axs, bars, titles):
ax.set(xticks=[],yticks=[],title=title)
divider = make_axes_locatable(ax)
ax_cb = divider.append_axes("right", size="5%", pad="2.5%")
py4DSTEM.visualize.add_colorbar_arg(ax_cb)
py4DSTEM.visualize.add_scalebar(ax,bar)
ab_fig.tight_layout()
ab_fig.canvas.resizable = False
ab_fig.canvas.header_visible = False
ab_fig.canvas.footer_visible = False
ab_fig.canvas.toolbar_visible = True
ab_fig.canvas.layout.width = '675px'
ab_fig.canvas.layout.height = '350px'
ab_fig.canvas.toolbar_position = 'bottom'def update_aberrated_plot(
semiangle_cutoff,
defocus_nm,
astigmatism_nm,
astigmatism_angle_deg,
coma_um,
coma_angle_deg,
):
""" """
ab_ctf, ab_fourier_probe, ab_complex_probe = return_probe_arrays(
semiangle_cutoff=semiangle_cutoff,
defocus=defocus_nm*10, # nm -> A
astigmatism=astigmatism_nm*10, # nm -> A
astigmatism_angle=np.deg2rad(astigmatism_angle_deg),
coma = coma_um*1e4, # um -> A
coma_angle = np.deg2rad(coma_angle_deg),
)
ab_rgb_fourier_probe = py4DSTEM.visualize.Complex2RGB(
ab_fourier_probe,
vmin=0,vmax=1,
)
ab_im_fourier.set_data(ab_rgb_fourier_probe)
ab_rgb_complex_probe = py4DSTEM.visualize.Complex2RGB(
ab_complex_probe,
vmin=0,vmax=1,
)
ab_im_complex.set_data(ab_rgb_complex_probe)
ab_fig.canvas.draw_idle()
return None
ab_layout = ipywidgets.Layout(width='330px',height='30px')
ab_semiangle_slider = ipywidgets.FloatSlider(
min=5,
max=50,
step=0.5,
value=20,
layout=ab_layout,
style=style,
description="semi-angle [mrad]",
)
ab_defocus_slider = ipywidgets.FloatSlider(
min=-15,
max=15,
step=0.5,
value=10,
layout=ab_layout,
style=style,
description="defocus [nm]",
)
ab_astigmatism_slider = ipywidgets.FloatSlider(
min=0,
max=10,
step=0.1,
value=5,
layout=ab_layout,
style=style,
description="astigmatism [nm]",
)
ab_coma_slider = ipywidgets.FloatSlider(
min=0,
max=2,
step=0.05,
value=0,
layout=ab_layout,
style=style,
description="coma [μm]",
)
ab_astigmatism_angle_slider = ipywidgets.FloatSlider(
min=-90,
max=90,
step=1,
value=45,
layout=ab_layout,
style=style,
description="astigmatism angle [°]",
)
ab_coma_angle_slider = ipywidgets.FloatSlider(
min=-180,
max=180,
step=1,
value=180,
layout=ab_layout,
style=style,
description="coma angle [°]",
)
ipywidgets.interactive_output(
update_aberrated_plot,
{
'semiangle_cutoff': ab_semiangle_slider,
'defocus_nm': ab_defocus_slider,
'astigmatism_nm': ab_astigmatism_slider,
'astigmatism_angle_deg': ab_astigmatism_angle_slider,
'coma_um': ab_coma_slider,
'coma_angle_deg': ab_coma_angle_slider
}
)
Nonedisplay(
ipywidgets.VBox(
[
ipywidgets.HBox([ab_semiangle_slider,ab_defocus_slider]),
ipywidgets.HBox([ab_astigmatism_slider,ab_astigmatism_angle_slider]),
ipywidgets.HBox([ab_coma_slider,ab_coma_angle_slider]),
ab_fig.canvas,
],
layout=ipywidgets.Layout(
align_items="center"
)
)
)VBox(children=(HBox(children=(FloatSlider(value=20.0, description='semi-angle [mrad]', layout=Layout(height='3…
