Contents
Detectors
# %matplotlib widget
import abtem
import ase
import numpy as np
import matplotlib.pyplot as plt
# import py4DSTEM
from scipy.ndimage import rotate
abtem.config.set({"dask.lazy":False});Atoms and Potential¶
srtio3_unit_cell = ase.io.read("data/SrTiO3.cif")
srtio3 = srtio3_unit_cell*(6,6,2)
gpts = (192,192)
scan_gpts = (32,32) # Double Nyquist later
energy = 200e3
semiangle_cutoff = 20potential = abtem.Potential(
srtio3,
gpts=gpts,
slice_thickness=srtio3.cell[2,2]/8,
projection='finite'
).build(
)probe_in_focus = abtem.Probe(
semiangle_cutoff=semiangle_cutoff,
energy=energy,
).match_grid(
potential
)Scans¶
custom_positions = srtio3_unit_cell.positions[:4,:2]
custom_scan = abtem.CustomScan(
positions=custom_positions
)line_scan = abtem.LineScan(
start=(0,0),
end=np.array(potential.extent)/6,
gpts=scan_gpts[0]*4
)grid_scan = abtem.GridScan(
start=(0,0),
end=np.array(potential.extent)/6,
gpts=scan_gpts,
)fig,ax = plt.subplots(figsize=(5,5))
abtem.show_atoms(
srtio3,
ax=ax
)
grid_scan.add_to_plot(ax,alpha=0.5,facecolor='b')
line_scan.add_to_axes(ax,color='k')
ax.scatter(custom_positions[:,1],custom_positions[:,0],c='magenta',marker='*',s=100);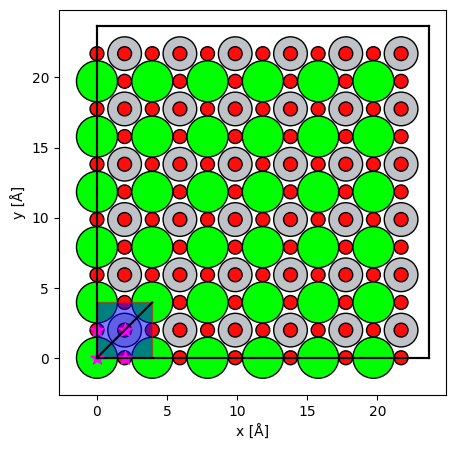
# exit_waves_custom = probe_in_focus.multislice(potential,scan=custom_scan)
exit_waves_line = probe_in_focus.multislice(potential,scan=line_scan)
exit_waves_grid = probe_in_focus.multislice(potential,scan=grid_scan)Monolithic Detectors¶
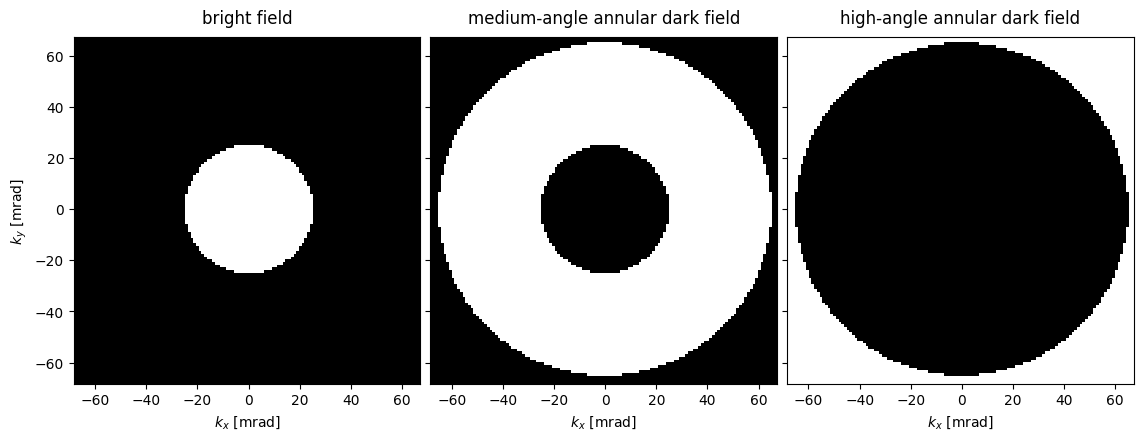
bright = abtem.AnnularDetector(inner=0, outer=25)
maadf = abtem.AnnularDetector(inner=25, outer=65)
haadf = abtem.AnnularDetector(inner=65, outer=100)bright_region = bright.get_detector_region(probe_in_focus)
maadf_region = maadf.get_detector_region(probe_in_focus)
haadf_region = haadf.get_detector_region(probe_in_focus)
labels = ("bright field", "medium-angle annular dark field", "high-angle annular dark field")
stacked_regions = abtem.stack(
(bright_region, maadf_region, haadf_region), labels
)
visualization = stacked_regions.show(
explode=True,units="mrad", figsize=(12,4.5),
cmap='gray'
)
Measurements¶
line_measurement_bright = bright.detect(exit_waves_line)
line_measurement_maadf = maadf.detect(exit_waves_line)
line_measurement_haadf = haadf.detect(exit_waves_line)line_measurements = abtem.stack(
[
line_measurement_bright,
line_measurement_maadf,
line_measurement_haadf
],
labels
)
visualization = line_measurements.show(explode=True,common_scale=False, figsize=(12, 3))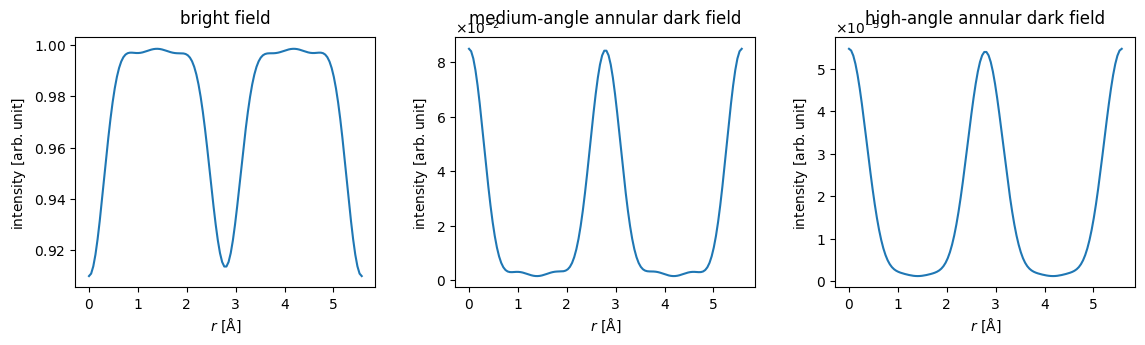
grid_measurement_bright = bright.detect(exit_waves_grid)
grid_measurement_maadf = maadf.detect(exit_waves_grid)
grid_measurement_haadf = haadf.detect(exit_waves_grid)grid_measurements = abtem.stack(
[
grid_measurement_bright,
grid_measurement_maadf,
grid_measurement_haadf
],
labels
)
visualization = grid_measurements.show(explode=True, cbar=True, figsize=(12, 3),cmap='magma')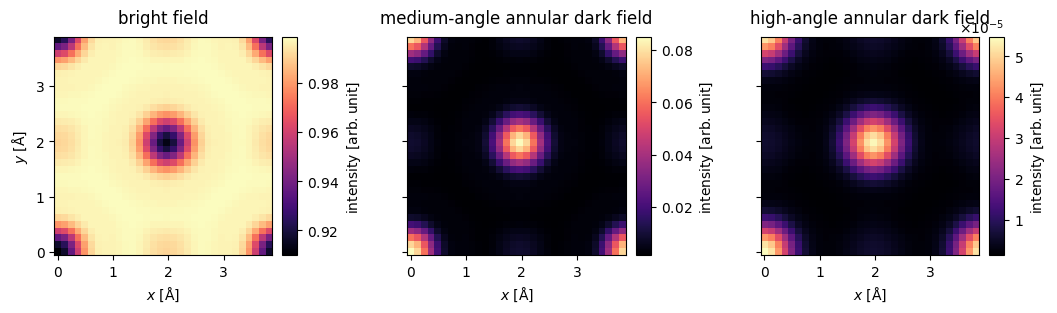
Pixelated Detectors¶
cropped_gpts = (gpts[0]//2,gpts[1]//2)
pixelated_detector = abtem.PixelatedDetector(max_angle=None)
grid_measurement_pixelated = pixelated_detector.detect(
exit_waves_grid
).crop(
gpts=(gpts[0]//2,gpts[1]//2)
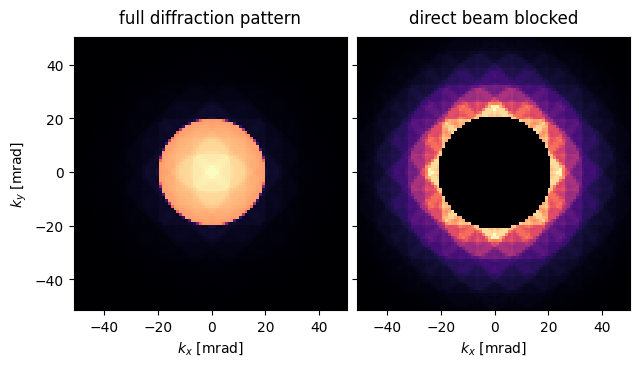
)abtem.stack(
[
grid_measurement_pixelated[0,0],
grid_measurement_pixelated[0,0].block_direct()
],
("full diffraction pattern","direct beam blocked")
).show(
explode=True,
units="mrad",
cmap='magma'
);
grid_measurement_pixelated_postprocessed = grid_measurement_pixelated.copy()
grid_measurement_pixelated_postprocessed._array = rotate(
grid_measurement_pixelated_postprocessed.array,
angle = 15, #deg
order=1,
reshape=False,
axes=(-1,-2)
).clip(0)
grid_measurement_pixelated_postprocessed = grid_measurement_pixelated_postprocessed.poisson_noise(
dose_per_area=1e5
)
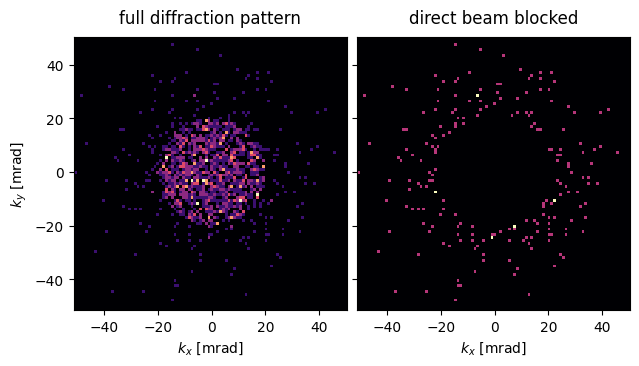
abtem.stack(
[
grid_measurement_pixelated_postprocessed[0,0],
grid_measurement_pixelated_postprocessed[0,0].block_direct(radius=semiangle_cutoff+0.25)
],
("full diffraction pattern","direct beam blocked")
).show(
explode=True,
units="mrad",
cmap='magma'
);
Export for use in 12.1.direct-ptychography.ipynb¶
# dc_in_focus = py4DSTEM.DataCube(grid_measurement_pixelated_postprocessed.array)
# dc_in_focus.calibration.set_Q_pixel_size(grid_measurement_pixelated_postprocessed.angular_sampling[0])
# dc_in_focus.calibration.set_R_pixel_size(grid_scan.sampling[0])
# dc_in_focus.calibration.set_Q_pixel_units('mrad')
# dc_in_focus.calibration.set_R_pixel_units('A')
# dc_in_focus.calibration# probe_defocus = abtem.Probe(
# semiangle_cutoff=semiangle_cutoff,
# energy=energy,
# C10=-100,
# ).match_grid(
# potential
# )
# grid_measurement_pixelated_postprocessed_defocus = probe_defocus.multislice(
# potential,
# scan=grid_scan,
# detectors=pixelated_detector
# ).crop(
# gpts=(gpts[0]//2,gpts[1]//2)
# )
# grid_measurement_pixelated_postprocessed_defocus._array = rotate(
# grid_measurement_pixelated_postprocessed_defocus.array,
# angle = 15, #deg
# order=1,
# reshape=False,
# axes=(-1,-2)
# ).clip(0)
# grid_measurement_pixelated_postprocessed_defocus = grid_measurement_pixelated_postprocessed_defocus.poisson_noise(
# dose_per_area=1e5
# )# probe_aberrated = abtem.Probe(
# semiangle_cutoff=semiangle_cutoff,
# energy=energy,
# C10=-100,C23=10000,phi23=np.deg2rad(27.5),
# ).match_grid(
# potential
# )
# grid_measurement_pixelated_postprocessed_aberrated = probe_aberrated.multislice(
# potential,
# scan=grid_scan,
# detectors=pixelated_detector
# ).crop(
# gpts=(gpts[0]//2,gpts[1]//2)
# )
# grid_measurement_pixelated_postprocessed_aberrated._array = rotate(
# grid_measurement_pixelated_postprocessed_aberrated.array,
# angle = 15, #deg
# order=1,
# reshape=False,
# axes=(-1,-2)
# ).clip(0)
# grid_measurement_pixelated_postprocessed_aberrated = grid_measurement_pixelated_postprocessed_aberrated.poisson_noise(
# dose_per_area=1e5
# )# dc_defocus = py4DSTEM.DataCube(grid_measurement_pixelated_postprocessed_defocus.array,calibration=dc_in_focus.calibration.copy())
# dc_aberrated = py4DSTEM.DataCube(grid_measurement_pixelated_postprocessed_aberrated.array,calibration=dc_in_focus.calibration.copy())# py4DSTEM.save("data/dpc_STO_simulation_in-focus_1e5.h5",dc_in_focus,mode='o')
# py4DSTEM.save("data/dpc_STO_simulation_defocus_1e5.h5",dc_defocus,mode='o')
# py4DSTEM.save("data/dpc_STO_simulation_aberrated_1e5.h5",dc_aberrated,mode='o')
