Contents
Converged Beam Blochwaves
import py4DSTEM
import abtem
import ase
import numpy as np
import matplotlib.pyplot as plt
from scipy.spatial.transform import Rotation
from matplotlib import cm
import matplotlib.colorsBlochwaves with py4DSTEM¶
Si3N4_crystal = ase.Atoms(
"Si6N8",
scaled_positions=[
(0.82495, 0.59387, 0.75),
(0.23108, 0.82495, 0.25),
(0.59387, 0.76892, 0.25),
(0.40614, 0.23108, 0.75),
(0.76892, 0.17505, 0.75),
(0.17505, 0.40613, 0.25),
(0.66667, 0.33334, 0.75),
(0.33334, 0.66667, 0.25),
(0.66986, 0.70066, 0.75),
(0.96920, 0.66986, 0.25),
(0.70066, 0.03081, 0.25),
(0.29934, 0.96919, 0.75),
(0.33015, 0.29934, 0.25),
(0.03081, 0.33014, 0.75),
],
cell=[7.6045, 7.6045, 2.9052, 90, 90, 120],
pbc=True
)
Si3N4_orthorhombic = abtem.orthogonalize_cell(Si3N4_crystal)
cell_thickness = Si3N4_orthorhombic.cell[2,2]
cell_thicknesses = np.arange(cell_thickness*50,cell_thickness*400,cell_thickness*100)g_max = 4
sg_max = 0.1
thermal_sigma = 0.1
energy = 300e3Si3N4_py4d = py4DSTEM.process.diffraction.Crystal(
positions=Si3N4_orthorhombic.get_scaled_positions(),
numbers=Si3N4_orthorhombic.numbers,
cell=Si3N4_orthorhombic.cell.array,
)
# Si3N4_py4d.calculate_structure_factors(
# k_max=g_max,
# tol_structure_factor=-1,
# )
Si3N4_py4d.calculate_dynamical_structure_factors(
energy,
"Lobato",
k_max=g_max,
thermal_sigma=thermal_sigma,
tol_structure_factor=-1,
)
ZA = np.array([0,0,1])
beams = Si3N4_py4d.generate_diffraction_pattern(
zone_axis_lattice=ZA,
sigma_excitation_error=sg_max,
)CBED Patterns¶
cbed_alpha_mrad = 1
cbed_reciprocal_pixelsize = 0.01cbeds = Si3N4_py4d.generate_CBED(
beams=beams,
thickness=cell_thicknesses,
alpha_mrad=cbed_alpha_mrad,
pixel_size_inv_A=cbed_reciprocal_pixelsize,
DP_size_inv_A=cbed_reciprocal_pixelsize*96,
zone_axis_lattice=ZA,
)100%|███████████████████████████████████████████████████████████████| 69/69 [00:23<00:00, 2.97it/s]
cbeds_array = np.array(cbeds).swapaxes(-1,-2)
_, vmin, vmax = py4DSTEM.visualize.return_scaled_histogram_ordering(cbeds_array[-1],vmax=0.975)
fig, axs = plt.subplots(1,4,figsize=(12,4))
py4DSTEM.show(
list(cbeds_array),
cmap='magma',
ticks=False,
vmin=vmin,vmax=vmax,intensity_range='absolute',
figax=(fig,axs),
scalebar={'position':'ur'},
origin='lower',
pixelsize=cbed_reciprocal_pixelsize,
pixelunits=r"$\AA^{-1}$"
)
for ax, thickness in zip(axs.ravel(),cell_thicknesses):
ax.texts[0].set_va('bottom')
ax.set_title(rf"{thickness:.3f} $\AA$")
fig.tight_layout()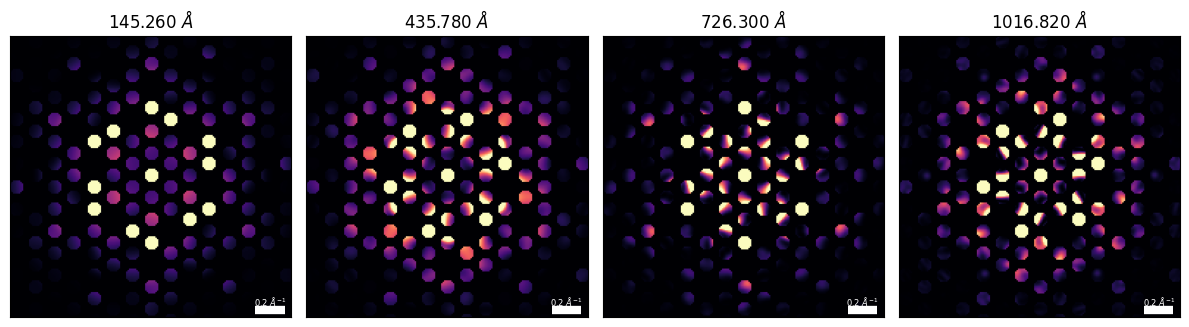
pacbed_alpha_mrad = 5
pacbed_reciprocal_pixelsize = 0.02pacbeds = Si3N4_py4d.generate_CBED(
beams=beams,
thickness=cell_thicknesses,
alpha_mrad=pacbed_alpha_mrad,
pixel_size_inv_A=pacbed_reciprocal_pixelsize,
DP_size_inv_A=pacbed_reciprocal_pixelsize*48,
zone_axis_lattice=ZA,
)100%|█████████████████████████████████████████████████████████████| 517/517 [02:53<00:00, 2.98it/s]
pacbeds_array = np.array(pacbeds).swapaxes(-1,-2)**0.25
_, vmin, vmax = py4DSTEM.visualize.return_scaled_histogram_ordering(pacbeds_array[-1],vmax=0.95)
fig, axs = plt.subplots(1,4,figsize=(12,4))
py4DSTEM.show(
list(pacbeds_array),
cmap='magma',
ticks=False,
vmin=vmin,vmax=vmax,intensity_range='absolute',
figax=(fig,axs),
scalebar={'position':'ur'},
origin='lower',
pixelsize=pacbed_reciprocal_pixelsize,
pixelunits=r"$\AA^{-1}$"
)
for ax, thickness in zip(axs.ravel(),cell_thicknesses):
ax.texts[0].set_va('bottom')
ax.set_title(rf"{thickness:.3f} $\AA$")
fig.tight_layout()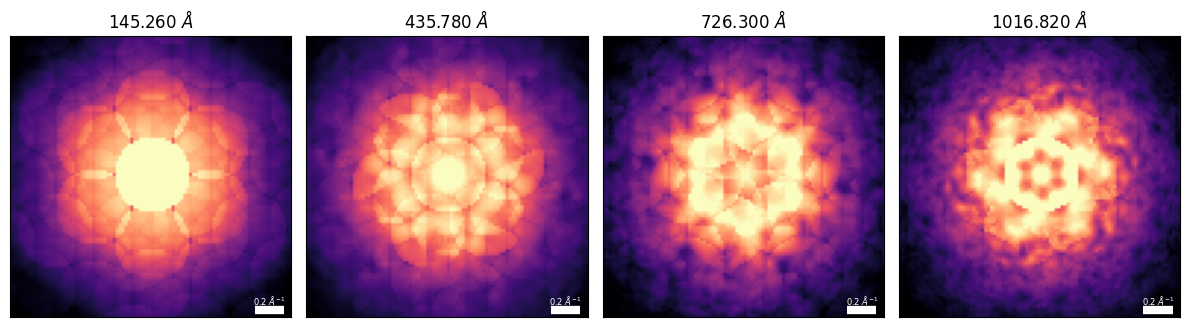
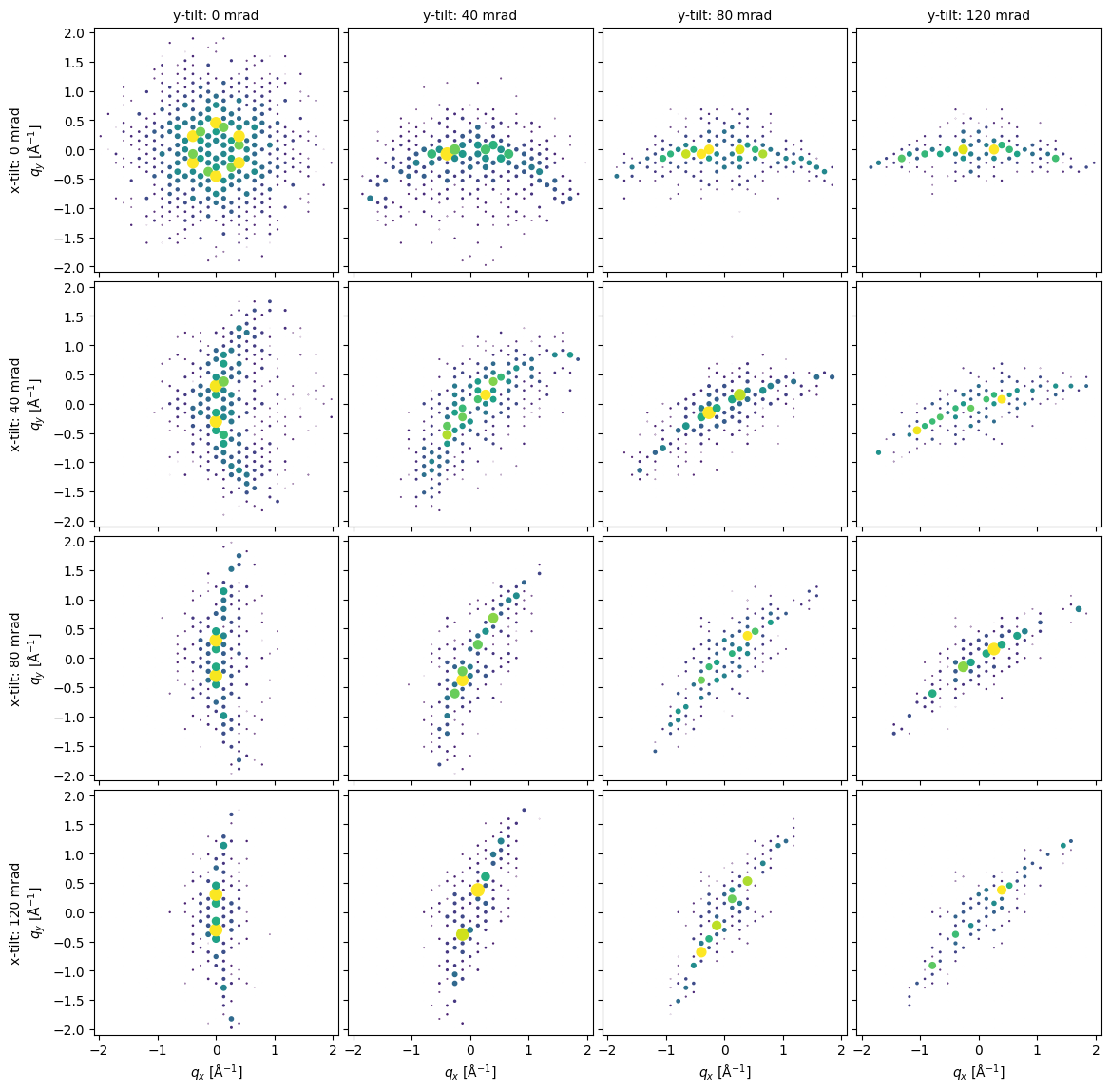
Tilted Diffraction Comparison¶
x_angles = np.linspace(0,120e-3,4)
y_angles = np.linspace(120e-3,0,4)
xy_angles = np.dstack(np.meshgrid(x_angles,y_angles,indexing='ij')).reshape((-1,2))
orientation_matrices = Rotation.from_euler('xy',xy_angles).as_matrix()
rotated_ZAs = Rotation.from_matrix(orientation_matrices).apply(ZA)fig = plt.figure(figsize=(12,12))
axes = abtem.visualize.axes_grid.AxesGrid(
fig=fig,
ncols=4,
nrows=4,
)._axes
y_indices = np.tile(np.arange(4),4)
x_indices = np.repeat(np.arange(4),4)
for za, ax, x_ind, y_ind in zip(rotated_ZAs,axes.ravel(),x_indices, y_indices):
dynamical_beams = Si3N4_py4d.generate_dynamical_diffraction_pattern(
beams=beams,
thickness=cell_thicknesses[0],
zone_axis_cartesian=za,
)
# block direct beam and clip low intensities
direct_index = np.where(
(dynamical_beams['h'] == 0) &
(dynamical_beams['k'] == 0) &
(dynamical_beams['l'] == 0)
)[0][0]
threshold = 2.5e-3
clipped_intensity = dynamical_beams['intensity'].copy()**0.5
clipped_intensity[direct_index] = threshold
clipped_intensity = clipped_intensity.clip(threshold) - threshold
dynamical_beams.data['intensity'] = clipped_intensity
py4DSTEM.process.diffraction.plot_diffraction_pattern(
dynamical_beams,
add_labels=False,
scale_markers=250,
input_fig_handle=(fig,(ax,))
)
ax.xaxis.tick_bottom()
ax.invert_yaxis()
# transpose x and y to match abtem
col = ax.collections[0]
col.set_offsets(np.flip(col.get_offsets(),axis=1))
# set colors
norm = matplotlib.colors.Normalize()
colors = cm.viridis(norm(clipped_intensity**0.5))
col.set_color(colors)
if x_ind == 0:
if y_ind == 0:
x_label = ax.get_xlabel()
y_label = ax.get_ylabel()
ax.set_ylabel(f"x-tilt: {x_angles[3-y_ind]*1e3:.0f} mrad\n"+x_label)
if y_ind == 3:
ax.set_title(f"y-tilt: {y_angles[3-x_ind]*1e3:.0f} mrad",fontsize=10)
if y_ind == 0:
ax.set_xlabel(y_label)