Contents
Blochwave Algorithm
import abtem
import abtem.bloch
import ase
import numpy as np
import matplotlib.pyplot as plt
from scipy.spatial.transform import Rotation
abtem.config.set({"dask.lazy":False});Si3N4_crystal = ase.Atoms(
"Si6N8",
scaled_positions=[
(0.82495, 0.59387, 0.75),
(0.23108, 0.82495, 0.25),
(0.59387, 0.76892, 0.25),
(0.40614, 0.23108, 0.75),
(0.76892, 0.17505, 0.75),
(0.17505, 0.40613, 0.25),
(0.66667, 0.33334, 0.75),
(0.33334, 0.66667, 0.25),
(0.66986, 0.70066, 0.75),
(0.96920, 0.66986, 0.25),
(0.70066, 0.03081, 0.25),
(0.29934, 0.96919, 0.75),
(0.33015, 0.29934, 0.25),
(0.03081, 0.33014, 0.75),
],
cell=[7.6045, 7.6045, 2.9052, 90, 90, 120],
pbc=True
)
Si3N4_orthorhombic = abtem.orthogonalize_cell(Si3N4_crystal)g_max = 4
sg_max = 0.1
thermal_sigma = 0.1
energy = 300e3
structure_factor = abtem.bloch.StructureFactor(
Si3N4_orthorhombic,
g_max=g_max,
thermal_sigma=thermal_sigma,
parametrization="lobato"
)
bloch_waves = abtem.bloch.BlochWaves(
structure_factor=structure_factor,
energy=energy,
sg_max=sg_max,
)
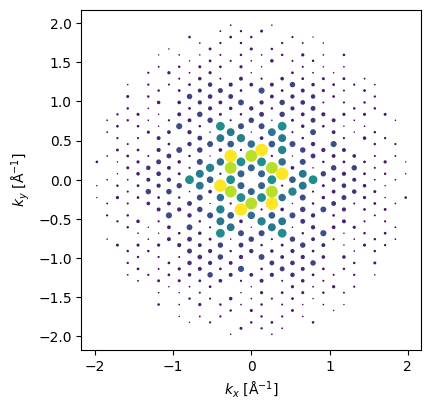
kinematical_diffraction_pattern = bloch_waves.get_kinematical_diffraction_pattern()kinematical_diffraction_pattern.block_direct(
).show(
scale=0.08,
power=0.5,
annotations=False,
figsize=(4,4)
);
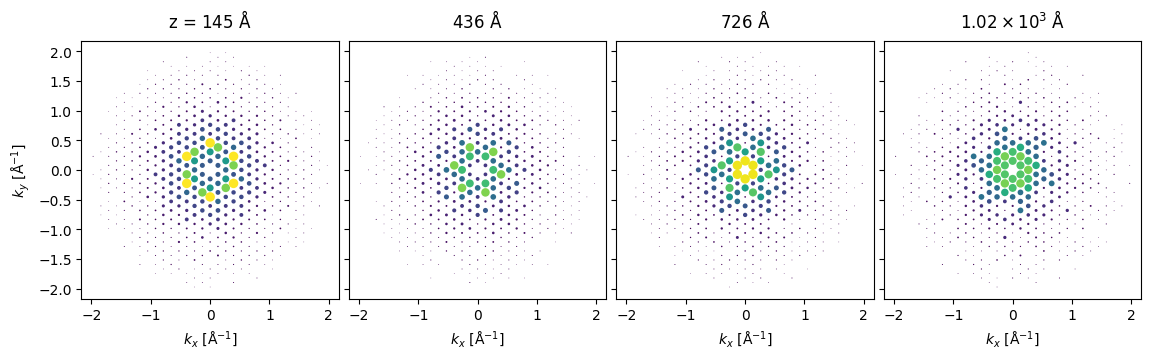
cell_thickness = Si3N4_orthorhombic.cell[2,2]
cell_thicknesses = np.arange(cell_thickness*50,cell_thickness*400,cell_thickness*100)bloch_waves.calculate_diffraction_patterns(
cell_thicknesses,
lazy=False,
).block_direct(
).show(
scale=0.08,
power=0.5,
explode=True,
annotations=False,
common_color_scale=True,
figsize=(12,4)
);
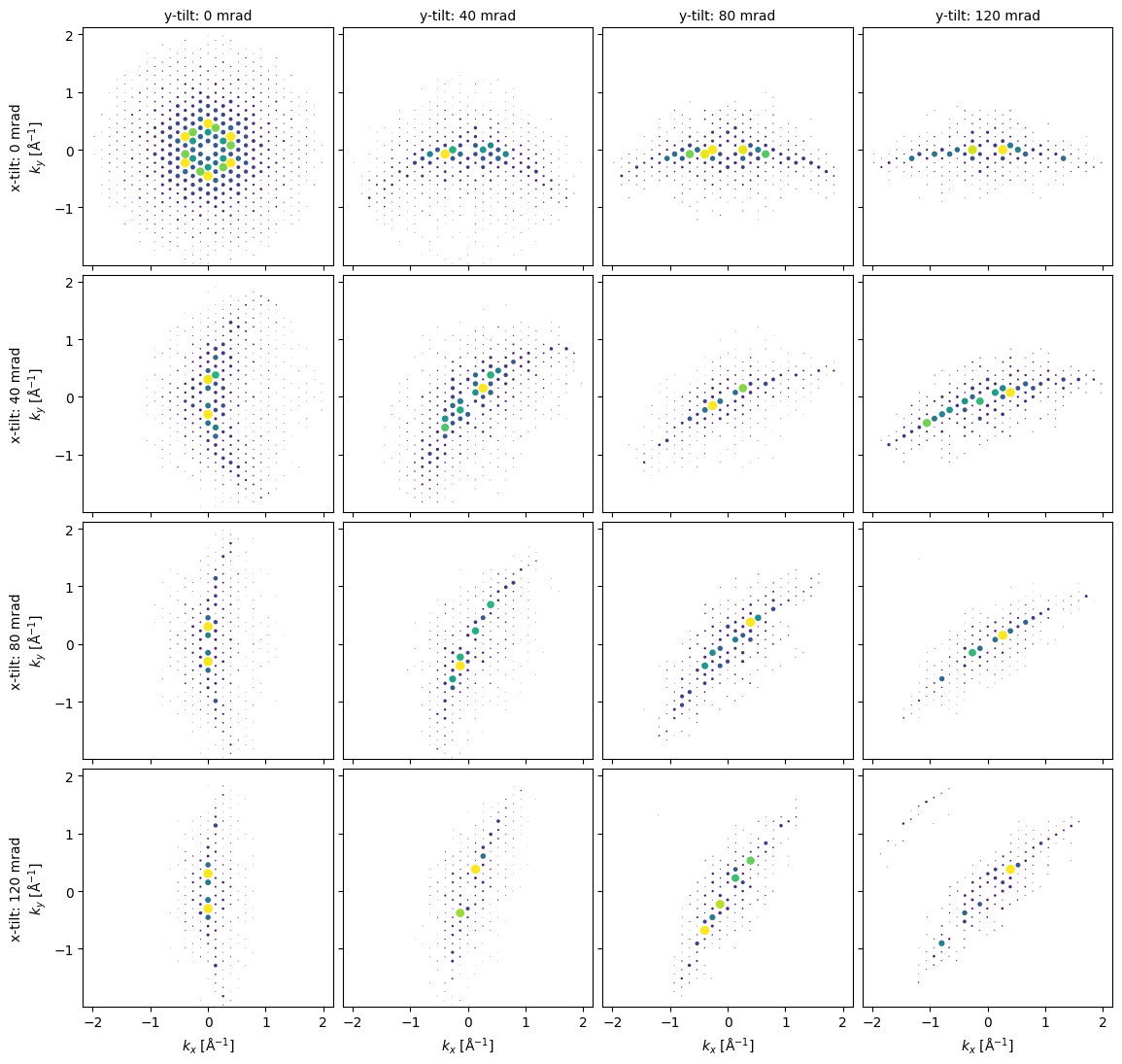
Tilted Crystal Electron Diffraction¶
x_angles = np.linspace(0,120e-3,4)
y_angles = np.linspace(120e-3,0,4)
xy_angles = np.dstack(np.meshgrid(x_angles,y_angles,indexing='ij')).reshape((-1,2))
orientation_matrices = Rotation.from_euler('xy',xy_angles).as_matrix()
bloch_matrices = [
abtem.bloch.BlochWaves(
structure_factor=structure_factor,
energy=energy,
sg_max=sg_max,
orientation_matrix=orientation,
) for orientation in orientation_matrices
]
dps = [
bloch.calculate_diffraction_patterns(
thicknesses=np.array([cell_thicknesses[0]]),
lazy=False,
).block_direct(
) for bloch in bloch_matrices
]
fig = plt.figure(figsize=(12,12))
axes = abtem.visualize.axes_grid.AxesGrid(
fig=fig,
ncols=4,
nrows=4,
)._axes
y_indices = np.tile(np.arange(4),4)
x_indices = np.repeat(np.arange(4),4)
for ax, dp, x_ind, y_ind in zip(axes.ravel(),dps,x_indices, y_indices):
dp.show(
scale=0.08,
power=0.5,
annotations=False,
ax=ax
)
if x_ind == 0:
label = ax.get_ylabel()
ax.set_ylabel(f"x-tilt: {x_angles[3-y_ind]*1e3:.0f} mrad\n"+label)
if y_ind == 3:
ax.set_title(f"y-tilt: {y_angles[3-x_ind]*1e3:.0f} mrad",fontsize=10)