Contents
Different Orientations Gold Widget
%matplotlib widget
import numpy as np
import ase
import abtem
import matplotlib.pyplot as plt
from matplotlib.gridspec import GridSpec
from scipy.spatial.transform import Rotation
import ipywidgets
import py4DSTEM
abtem.config.set({"dask.lazy":False});def inside_cell(positions,cell_dim):
lx,ly,lz = cell_dim/2
x,y,z = positions.T
return np.logical_and.reduce(
(
x > -lx,
x < lx,
y > -ly,
y < ly,
z > -lz,
z < lz,
)
)
def correct_side_of_plane(positions,plane_vec,plane_pos,side="left"):
expression = plane_vec @ (positions - plane_pos).T
if side == "left":
indices = expression < -1
elif side == "right":
indices = expression > 1
else:
raise ValueError()
return indices# gold = ase.build.bulk("Au",cubic=True,a=4)
gold_unit_cell = ase.Atoms(
symbols="Au4",
scaled_positions=[
[0.25,0.25,0.25],
[0.75,0.75,0.25],
[0.75,0.25,0.75],
[0.25,0.75,0.75]
],
cell=[4,4,4],
pbc=True
)
gold = gold_unit_cell * (21,21,21)
gold.translate(-10*4) # center cell_dim = np.array([150,40,40])
grain_angles = np.array(
[
[0,0,76],
[0,45,110],
[45,-54.73,22]
]
)
grain_centroids = np.array(
[
[-50,-4,-8],
[0,4,10],
[50,0,2]
]
)
grain_vectors = np.diff(grain_centroids,axis=0).astype(np.float64)
grain_vectors /= np.linalg.norm(grain_vectors,axis=1)[:,None]
grain_midpoints = grain_centroids[[[0,1],[1,2]]].mean(1)# Euler to angle/axis
rot_vecs = Rotation.from_euler(
"zxz",
grain_angles,
degrees=True
).as_rotvec()
norms = np.linalg.norm(rot_vecs,axis=1)
angles = np.rad2deg(norms)
axes = rot_vecs / norms[:,None]grains = []
for angle, axis, centroid in zip(angles,axes,grain_centroids):
grain = gold.copy()
grain.rotate(angle,axis)
grain.translate(centroid)
grains.append(grain)
grain_left, grain_middle, grain_right = grains
grain_left = grain_left[inside_cell(
grain_left.positions,
cell_dim
)]
grain_left = grain_left[correct_side_of_plane(
grain_left.positions,
grain_vectors[0],
grain_midpoints[0],
"left"
)]
grain_middle = grain_middle[inside_cell(
grain_middle.positions,
cell_dim
)]
grain_middle = grain_middle[correct_side_of_plane(
grain_middle.positions,
grain_vectors[0],
grain_midpoints[0],
"right"
)]
grain_middle = grain_middle[correct_side_of_plane(
grain_middle.positions,
grain_vectors[1],
grain_midpoints[1],
"left"
)]
grain_right = grain_right[inside_cell(
grain_right.positions,
cell_dim
)]
grain_right = grain_right[correct_side_of_plane(
grain_right.positions,
grain_vectors[1],
grain_midpoints[1],
"right"
)]n_left = grain_left.positions.shape[0]
n_middle = grain_middle.positions.shape[0]
n_right = grain_right.positions.shape[0]
grain = grain_left + grain_middle + grain_right
grain.translate(cell_dim/2)
grain.cell = cell_dim
grain_visualize = grain.copy()
grain_visualize.numbers[:n_left] += 1
grain_visualize.numbers[-n_right:] += 2potential = abtem.Potential(
grain,
sampling=0.2,
slice_thickness=cell_dim[2],
).build(
)energy = 300e3
semiangle_cutoff = 3
probe = abtem.Probe(
semiangle_cutoff=semiangle_cutoff,
energy=energy,
).match_grid(
potential
)detector = abtem.PixelatedDetector(max_angle=None)array_to_mutate = [
np.array([25,20])
]pos = np.array(array_to_mutate)
custom_scan = abtem.CustomScan(pos)dp = probe.scan(
potential,
custom_scan,
detector
).crop(
max_angle=probe.cutoff_angles[0]
).block_direct(
)dpi = 72
with plt.ioff():
fig = plt.figure(figsize=(675/dpi,200/dpi),dpi=dpi)
gs = GridSpec(1, 2, figure=fig,width_ratios=[4,1])
ax1 = fig.add_subplot(gs[0])
ax2 = fig.add_subplot(gs[1])
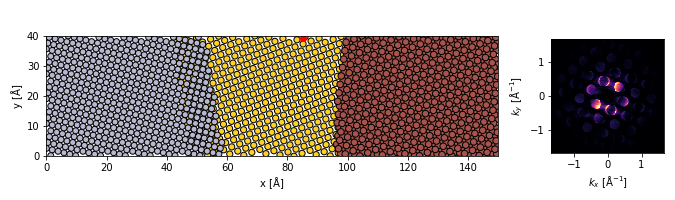
abtem.show_atoms(grain_visualize,plane='xy',tight_limits=True,ax=ax1)
scatter = ax1.scatter(
pos[0,0],
pos[0,1],
color='r'
)
dp.show(
cmap='magma',
ax=ax2
)
dp_im = ax2.images[0]
gs.tight_layout(fig)
fig.canvas.resizable = False
fig.canvas.header_visible = False
fig.canvas.footer_visible = False
fig.canvas.toolbar_visible = True
fig.canvas.layout.width = '680px'
fig.canvas.layout.height = "235px"
fig.canvas.toolbar_position = 'bottom'
def update_plots():
""" """
pos = np.array(array_to_mutate)
custom_scan = abtem.CustomScan(pos)
dp = probe.scan(
potential,
custom_scan,
detector
).crop(
max_angle=probe.cutoff_angles[0]
).block_direct(
)
dp_im.set_data(dp.array)
scatter.set_offsets(pos)
fig.canvas.draw_idle()
return None
def onmove(event):
""" """
pos = np.array([event.xdata,event.ydata])
if pos[0] is not None:
array_to_mutate[0] = pos
update_plots()
return None
cid = fig.canvas.mpl_connect('motion_notify_event',onmove)
Nonefig.canvas