Direct Ptychography Tutorial 01
This is the first tutorial notebook in the direct ptychography series.
In this tutorial notebook we will cover:
- Complex probe overlap functions
- Single side band (SSB) reconstructions
Downloads¶
This tutorial uses the following datasets:
- Georgios Varnavides (gvarnavides@berkeley
.edu) - Stephanie Ribet (sribet@lbl.gov)
- Colin Ophus (cophus@stanford.edu)
Last updated: 2025 Feb 01
Introduction¶
Direct ptychography techniques are a class of phase retrieval methods which attempts to reconstruct the scattering potential of an electron-transparent sample by deconvolving the effect of the incident illumination (probe aperture and aberrations).
%pip install py4DSTEM > /dev/null 2>&1import numpy as np
import py4DSTEM
import matplotlib.pyplot as plt
print(py4DSTEM.__version__)
%matplotlib inlinecupyx.jit.rawkernel is experimental. The interface can change in the future.
0.14.18
# Get the 4DSTEM data
py4DSTEM.io.gdrive_download(
id_ = 'https://drive.google.com/uc?id=1Ml1Ga4-U0ZcTJxq0LVyTJDIVfnBuyd81',
destination = '/content/',
filename = 'dpc_STO_simulation.h5',
overwrite=True
)Downloading...
From: https://drive.google.com/uc?id=1Ml1Ga4-U0ZcTJxq0LVyTJDIVfnBuyd81
To: /content/dpc_STO_simulation.h5
100%|██████████| 75.5M/75.5M [00:00<00:00, 114MB/s]
file_path = '/content/'
file_data = file_path + 'dpc_STO_simulation.h5'
dataset = py4DSTEM.read(file_data)
datasetDataCube( A 4-dimensional array of shape (32, 32, 96, 96) called 'datacube',
with dimensions:
Rx = [0.0,0.12328531250000001,0.24657062500000002,...] A
Ry = [0.0,0.12328531250000001,0.24657062500000002,...] A
Qx = [0.0,1.0595062826244177,2.1190125652488354,...] mrad
Qy = [0.0,1.0595062826244177,2.1190125652488354,...] mrad
)Inspecting the datacube¶
First, let’s inspect the dataset, , with real-space dimensions and dimensions.
By selecting a pair of positions, we can visualize an example noisy diffraction pattern, and by summing of we can boost the signal-to-noise ratio and get an image of the bright-field disk:
py4DSTEM.show(
[
dataset[0,0],
dataset.data.mean((0,1))
],
ticks=False,
scalebar=True,
pixelsize=dataset.calibration.Q_pixel_size,
pixelunits=dataset.calibration.Q_pixel_units,
axsize=(4,4),
title=["example diffraction pattern","mean diffraction pattern"]
)
We can also take the Fourier-transform along the real-space dimensions, to obtain another datacube , where are the real-space scan frequencies. Not the dataset will now be complex-valued, so we plot them using show_complex:
dataset_real_space_FFT = np.fft.fft2(np.fft.ifftshift(dataset.data,axes=(-1,-2)),axes=(0,1))py4DSTEM.show_complex(
[
np.fft.fftshift(dataset_real_space_FFT[0,2]),
np.fft.fftshift(dataset_real_space_FFT[0,4])
],
ticks=False,
axsize=(4,4),
title=["low frequency overlap trotter","high frequency overlap trotter"]
)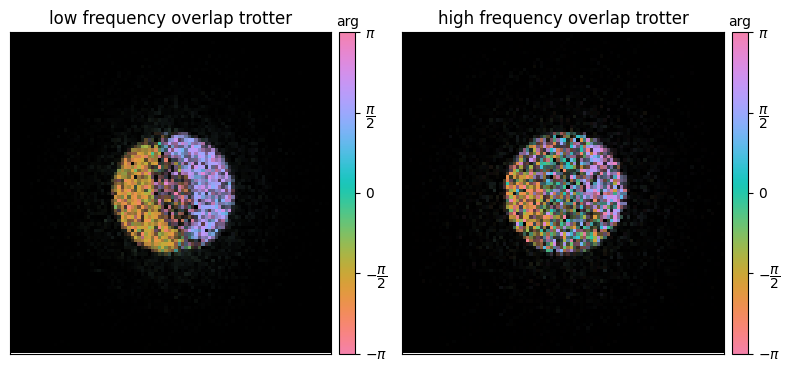
Visualized in this way, the dataset holds a wealth of information:
- The amplitude of the arrays above for each spatial frequency arises from the overlap between the bright-field aperture and two apertures centered at .
- At high spatial frequencies (right panel), this forms two regions of “double-overlap”: and
- At low spatial frequencies (left panel), the “double-overlap” regions can themselves overlap, leading to the “triple-overlap” region
- At zero-defocus (as above), the “triple-overlap” region cancels out and thus holds no information
- The similarity of the low-spatial frequency probe overlap regions, gave rise to the colloquialism we’ll also adopt, calling them trotters
- More importantly, note that the phase of each “trotter” is constant (for zero-defocus) and π-shifted from the other
- It’s precisely this information we’ll use to extract the sample phase by deconvolving the effects of the probe aperture (and aberrations)
- The relative rotation of the dataset (here -15°), can also be visualized as the offset of the perpendicular axis between the two trotters
Modeling probe convolution¶
If we know our probe aperture (convergence semi-angle) and our probe aberrations well, we can model the effect on the probe in the absence of a sample.
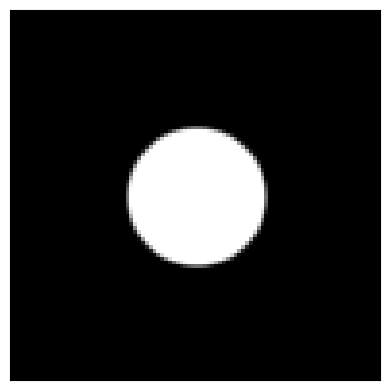
First, let’s model our bright-field probe aperture:
from py4DSTEM.process.phase.utils import ComplexProbe
energy = 200e3 # V
wavelength = py4DSTEM.process.utils.electron_wavelength_angstrom(energy)
semiangle_cutoff = 20 # mrad
gpts = dataset.Qshape
alpha_max = dataset.Q_pixel_size * gpts[0] / 2
k_max = alpha_max * 1e-3 / wavelength
sampling = (1 / k_max / 2, 1 / k_max / 2)
Kx, Ky = tuple(np.fft.fftfreq(n,s) for n,s in zip(gpts,sampling))
bright_field_probe = ComplexProbe(
energy=energy,
gpts=gpts,
sampling=sampling,
semiangle_cutoff=semiangle_cutoff,
force_spatial_frequencies=(Kx,Ky)
)
bright_field_alpha, bright_field_phi = bright_field_probe.get_scattering_angles()
bright_field_aperture = bright_field_probe.evaluate_aperture(bright_field_alpha,bright_field_phi)
py4DSTEM.show(
np.fft.fftshift(bright_field_aperture),
ticks=False,
figsize=(4,4),
)
We know (see dpc_01.ipynb tutorial for how to obtain this from the dataset directly) that our dataset is rotated by 15°.
Equivalently, we can rotate the probe-positions (and thus the grid in which we evaluate our spatial frequencies )
scan_gpts = dataset.Rshape
scan_sampling = (dataset.calibration.R_pixel_size,)*2
rotation_angle = np.deg2rad(-15)
ct = np.cos(-rotation_angle)
st = np.sin(-rotation_angle)
Qx, Qy = tuple(np.fft.fftfreq(n,s) for n,s in zip(scan_gpts,scan_sampling))
Qx, Qy = np.meshgrid(Qx,Qy, indexing='ij')
Qx, Qy = Qx * ct - Qy * st, Qy * ct + Qx * st # rotateIf we now pick one low-spatial frequency, we can form the two shifted apertures like above:
shifted_plus_probe = ComplexProbe(
energy=energy,
gpts=gpts,
sampling=sampling,
semiangle_cutoff=semiangle_cutoff,
force_spatial_frequencies=(Kx+Qx[0,2],Ky+Qy[0,2])
)
shifted_plus_alpha, shifted_plus_phi = shifted_plus_probe.get_scattering_angles()
shifted_plus_aperture = shifted_plus_probe.evaluate_aperture(shifted_plus_alpha,shifted_plus_phi)
shifted_minus_probe = ComplexProbe(
energy=energy,
gpts=gpts,
sampling=sampling,
semiangle_cutoff=semiangle_cutoff,
force_spatial_frequencies=(Kx-Qx[0,2],Ky-Qy[0,2])
)
shifted_minus_alpha, shifted_minus_phi = shifted_minus_probe.get_scattering_angles()
shifted_minus_aperture = shifted_minus_probe.evaluate_aperture(shifted_minus_alpha,shifted_minus_phi)
double_overlap_plus = np.logical_and(bright_field_aperture,shifted_plus_aperture)
double_overlap_minus = np.logical_and(bright_field_aperture,shifted_minus_aperture)
double_minus_triple_overlap_plus = np.logical_and(double_overlap_plus,1-double_overlap_minus)
double_minus_triple_overlap_minus = np.logical_and(double_overlap_minus,1-double_overlap_plus)
fig, axs = plt.subplots(1,3,figsize=(9,3))
py4DSTEM.show(
[
np.fft.fftshift(bright_field_aperture),
np.fft.fftshift(shifted_plus_aperture),
np.fft.fftshift(shifted_minus_aperture),
],
ticks=False,
figax=(fig,axs[0]),
combine_images=True,
title="superimposed apertures"
)
py4DSTEM.show(
[
np.fft.fftshift(double_overlap_plus),
np.fft.fftshift(double_overlap_minus),
],
ticks=False,
figax=(fig,axs[1]),
combine_images=True,
title="superimposed double overlap"
)
py4DSTEM.show(
[
np.fft.fftshift(double_minus_triple_overlap_plus),
np.fft.fftshift(double_minus_triple_overlap_minus),
],
ticks=False,
figax=(fig,axs[2]),
combine_images=True,
title="double - triple overlap"
)
fig.tight_layout()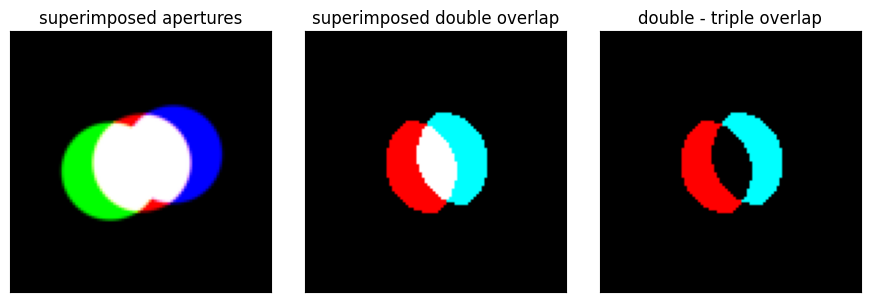
Deconvolving probe from dataset¶
We can use on of these double-triple overlap masks, hence the name single side band (SSB), to sum the phase of each trotter to obtain a single complex-value for each spatial frequency .
Inverse Fourier transforming that gives us our reconstructed object
reconstructed_object_fft = np.zeros(dataset.Rshape,dtype=np.complex64)
for sx, sy in py4DSTEM.tqdmnd(dataset.R_Nx,dataset.R_Ny):
shifted_plus_probe = ComplexProbe(
energy=energy,
gpts=gpts,
sampling=sampling,
semiangle_cutoff=semiangle_cutoff,
force_spatial_frequencies=(Kx+Qx[sx,sy],Ky+Qy[sx,sy])
)
shifted_plus_alpha, shifted_plus_phi = shifted_plus_probe.get_scattering_angles()
shifted_plus_aperture = shifted_plus_probe.evaluate_aperture(shifted_plus_alpha,shifted_plus_phi)
shifted_minus_probe = ComplexProbe(
energy=energy,
gpts=gpts,
sampling=sampling,
semiangle_cutoff=semiangle_cutoff,
force_spatial_frequencies=(Kx-Qx[sx,sy],Ky-Qy[sx,sy])
)
shifted_minus_alpha, shifted_minus_phi = shifted_minus_probe.get_scattering_angles()
shifted_minus_aperture = shifted_minus_probe.evaluate_aperture(shifted_minus_alpha,shifted_minus_phi)
double_overlap_plus = np.logical_and(bright_field_aperture,shifted_plus_aperture)
double_overlap_minus = np.logical_and(bright_field_aperture,shifted_minus_aperture)
double_minus_triple_overlap_minus = np.logical_and(double_overlap_minus,1-double_overlap_plus)
G = dataset_real_space_FFT[sx,sy]
reconstructed_object_fft[sx,sy] = G[double_minus_triple_overlap_minus].sum() * 2
# set scale
reconstructed_object_fft[0,0] = np.abs(dataset_real_space_FFT[0,0]).sum()
mean_intensity = dataset.data.sum((-1,-2)).mean()
reconstructed_object = np.fft.ifft2(reconstructed_object_fft) / mean_intensity100%|██████████| 1024/1024 [00:00<00:00, 1187.67it/s]
Let’s plot our reconstructed object -- note we recover both the amplitude and phase of the object:
fig, axs = plt.subplots(1,2,figsize=(8,4))
for ax, arr, cmap, title in zip(
axs,
(np.abs(reconstructed_object),np.angle(reconstructed_object)),
('gray','magma'),
('reconstructed amplitude','reconstructed phase')
):
py4DSTEM.show(
np.tile(arr,(2,2)),
figax=(fig,ax),
cmap=cmap,
ticks=False,
show_cbar=True,
title=title,
)
fig.tight_layout()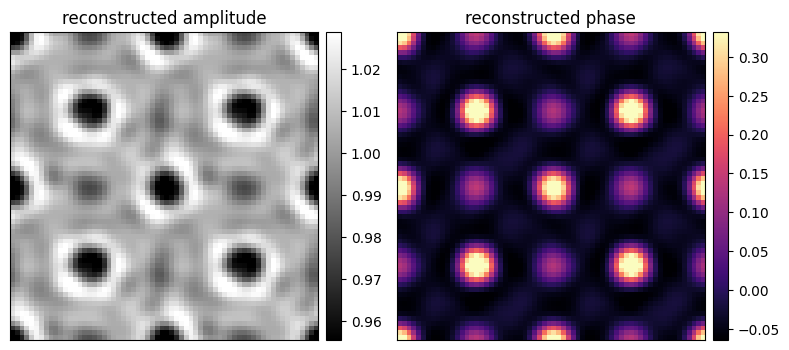
py4DSTEM implementation¶
Now that we understand what direct ptychography does, let’s look at the py4DSTEM.process.phase.SSB implementation:
ssb = py4DSTEM.process.phase.SSB(
energy=energy,
datacube=dataset,
semiangle_cutoff=semiangle_cutoff,
verbose=True,
).preprocess(
plot_center_of_mass=False,
plot_rotation=False,
force_com_rotation=-15,
vectorized_com_calculation=False,
crop_around_bf_disk=True,
bf_disk_padding_px = 2,
)Calculating center of mass: 100%|██████████| 1024/1024 [00:00<00:00, 34287.60probe position/s]
Best fit rotation forced to -15 degrees.
Normalizing amplitudes: 100%|██████████| 1024/1024 [00:01<00:00, 851.39probe position/s]
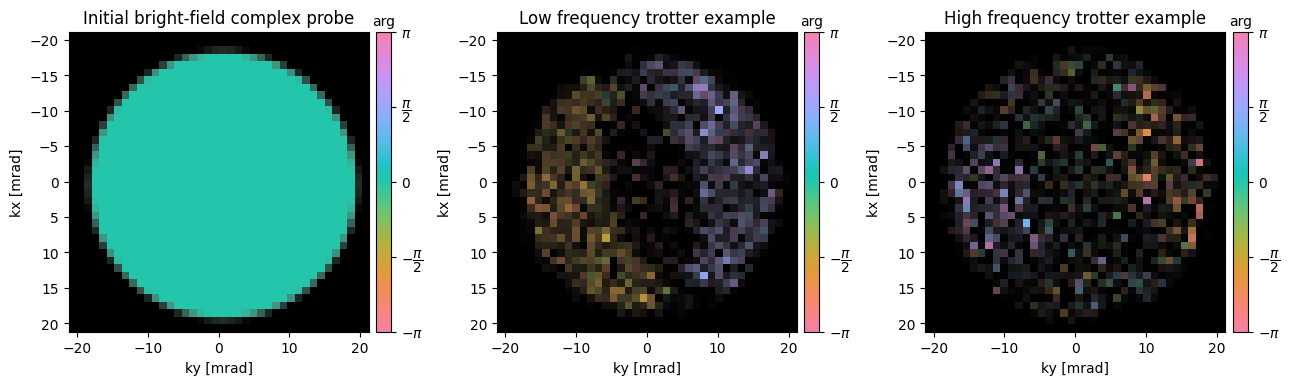
First, notice that since all the information in the double minus triple overlap region is (by construction) contained inside the BF-disk, we can crop our dataset, to reduce computation time.
As above, we can use one of the trotters to reconsruct the dataset by setting phase_compensation=False during reconstruction.
ssb = ssb.reconstruct(
phase_compensation=False,
progress_bar=True,
).visualize(
)
reconstructed_object_ssb = ssb.object.copy()100%|██████████| 1024/1024 [00:00<00:00, 1671.13it/s]
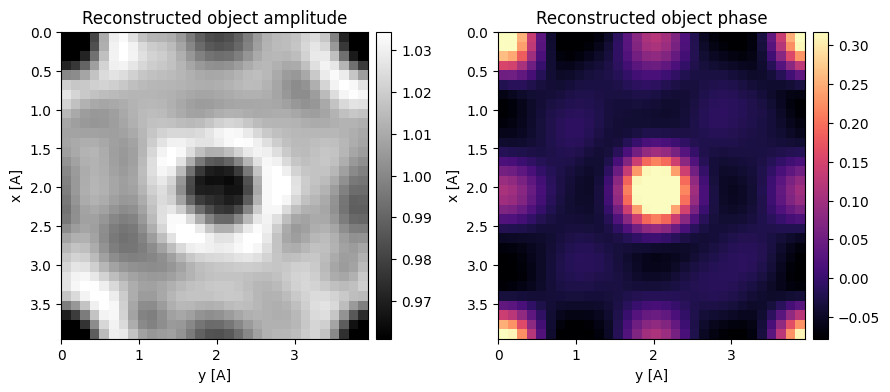
We can in-fact use both the trotters, by properly compensating their relative phase by setting phase_compensation=True (the default).
This uses the formalism described in [Yang, H. et al. (2016), Ultramic, 171, 117-125] and will further be useful in the case of aberrated probe datasets, see the direct_ptychography_02.ipynb tutorial.
ssb = ssb.reconstruct(
phase_compensation=True,
progress_bar=True,
).visualize(
)
reconstructed_object_compensated_ssb = ssb.object.copy()100%|██████████| 1024/1024 [00:00<00:00, 1262.54it/s]
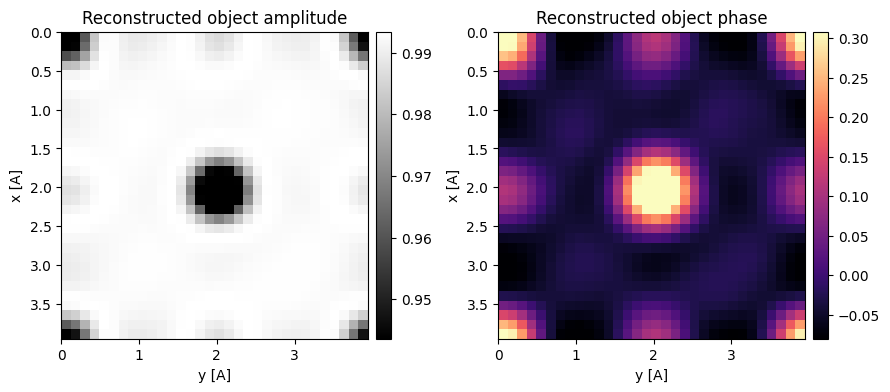
Notice that the phase of the two reconstructions are quite comparable, but the latter more accuralely regularized the amplitude term.
fig, axs = plt.subplots(1,3,figsize=(12,4))
for ax, arr, title in zip(
axs,
(np.angle(reconstructed_object),np.angle(reconstructed_object_ssb),np.angle(reconstructed_object_compensated_ssb)),
('reconstructed phase (SSB)','reconstructed phase (py4DSTEM SSB)','reconstructed phase (py4DSTEM SSB*)')
):
py4DSTEM.show(
np.tile(arr,(2,2)),
figax=(fig,ax),
cmap='turbo',
ticks=False,
title=title,
)
fig.tight_layout()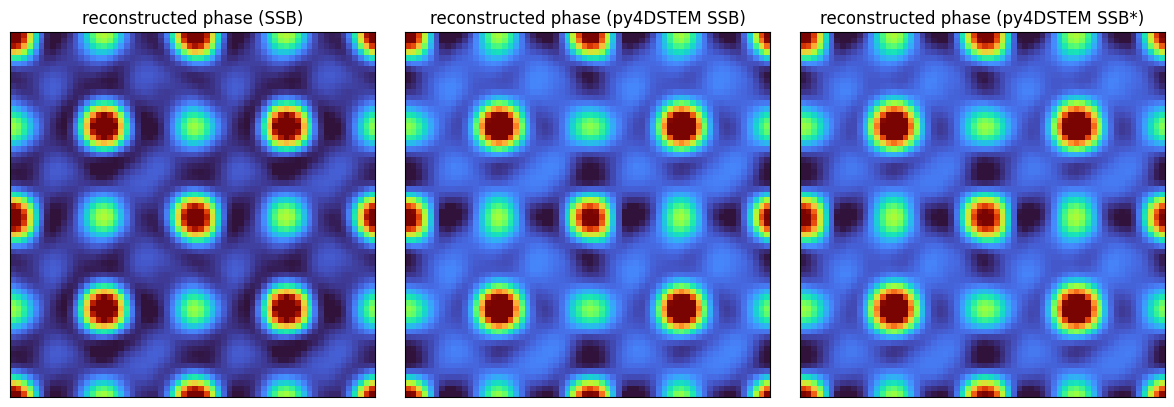
Acknowledgments¶
This tutorial was created by the py4DSTEM phase_contrast team:
- Yang, H., Ercius, P., Nellist, P. D., & Ophus, C. (2016). Enhanced phase contrast transfer using ptychography combined with a pre-specimen phase plate in a scanning transmission electron microscope. Ultramicroscopy, 171, 117–125. 10.1016/j.ultramic.2016.09.002

