Contents
Partial Molar Entropy
import numpy as np
import matplotlib.pyplot as plt
import plotly.graph_objects as goalpha = np.linspace(0.0, 1.0, num=1001, endpoint=False)
alpha = alpha[1:]
normalized_entropy = - 1 / alpha * (alpha * np.log(alpha)
+ (1 - alpha) * np.log(1 - alpha))
partial_molar_entropy = - np.log(alpha / (1 - alpha))fig, ax = plt.subplots(figsize=(6, 5))
ax.axhline(y=0, lw=1, c="grey", alpha=0.3)
ax.plot(alpha, normalized_entropy, c="r", lw=2,
label=r"$\Delta \overline{S}_{\mathrm{mix}, \mathrm{CO}_2}$")
ax.plot(alpha, partial_molar_entropy, c="g", lw=2,
label=r"$\Delta s_{\mathrm{mix}, \mathrm{CO}_2}$")
ax.set_xlabel(r"$\alpha_{\mathrm{CO}_2}$", fontsize=15)
ax.set_ylabel(r"$\frac{\Delta S_{\mathrm{mix}}}{n_{\mathrm{total}}\cdot R}$",
fontsize=15, labelpad=20, rotation=0,)
ax.set_xlim((0, 1))
ax.set_ylim((np.min(partial_molar_entropy), np.max(partial_molar_entropy)))
ax.set_yticks(np.atleast_1d(0))
ax.legend()
plt.tight_layout()
plt.show()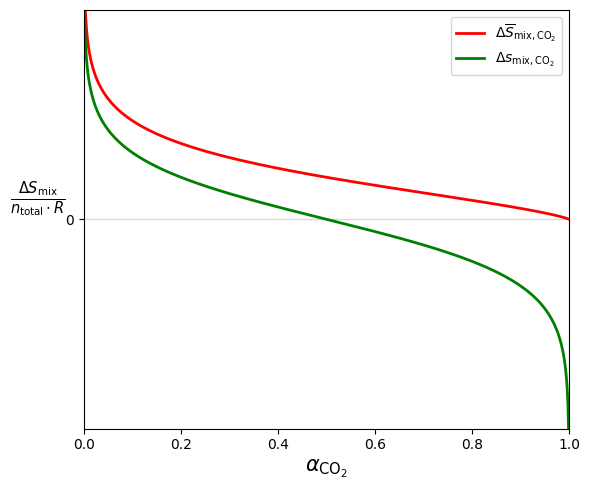
# --- Define the function and derivative ---
def y_func(x):
return -(x*np.log(x) + (1-x)*np.log(1-x))
def dy_dx(x):
return -np.log(x/(1.0 - x))
# --- Sample points for the main red curve ---
x_vals = np.linspace(0.001, 0.999, 200)
y_vals = y_func(x_vals)
# --- Choose slider positions for x in (0,1) ---
slider_xs = np.linspace(0.001, 0.999, 30)
##############################################
# 1) Initial value so everything shows at start
##############################################
x_init = slider_xs[0]
y_init = y_func(x_init)
slope_init = dy_dx(x_init)
xtan_init = np.linspace(0, 1, 50)
ytan_init = y_init + slope_init*(xtan_init - x_init)
xspec_init = [0, x_init]
yspec_init = [0, y_init]
##############################################
# 2) Base figure with 4 traces
##############################################
fig = go.Figure(
data=[
# Trace 0: main curve (red)
go.Scatter(
x=x_vals,
y=y_vals,
mode='lines',
line=dict(color='red'),
name='Function'
),
# Trace 1: black dot
go.Scatter(
x=[x_init],
y=[y_init],
mode='markers',
marker=dict(color='black', size=8),
name='Current Point'
),
# Trace 2: tangent line
go.Scatter(
x=xtan_init,
y=ytan_init,
mode='lines',
line=dict(color='black', dash='dash'),
name='Partial Molar (Tangent)'
),
# Trace 3: specific line
go.Scatter(
x=xspec_init,
y=yspec_init,
mode='lines',
line=dict(color='blue', dash='dash'),
name='Specific'
),
],
layout=go.Layout(
# Remove/empty the main title
title="",
xaxis=dict(range=[0,1], title='X'),
yaxis=dict(range=[0,0.7], title='Y'),
# Make the legend horizontal and place it at the top
legend=dict(
orientation="h",
yanchor="bottom",
y=1.02,
xanchor="center",
x=0.5
)
)
)
##############################################
# 3) Frames for animation
##############################################
frames = []
steps = []
for xv in slider_xs:
yv = y_func(xv)
slope = dy_dx(xv)
xtan = np.linspace(0, 1, 50)
ytan = yv + slope*(xtan - xv)
xspec = [0, xv]
yspec = [0, yv]
frame_name = f"x={xv:.2f}"
frame_data = [
{}, # keep the red curve
dict(x=[xv], y=[yv]), # black dot
dict(x=xtan, y=ytan), # tangent
dict(x=xspec, y=yspec), # specific line
]
frames.append(go.Frame(data=frame_data, name=frame_name))
steps.append(
dict(
method="animate",
label=frame_name,
args=[
[frame_name],
dict(frame=dict(duration=0, redraw=True),
mode="immediate",
transition=dict(duration=0))
]
)
)
fig.frames = frames
##############################################
# 4) Slider & "Play" button
##############################################
fig.update_layout(
template="simple_white",
width=700,
height=500,
yaxis=dict(range=[0, 1]),
sliders=[
dict(
active=0,
steps=steps,
x=0,
y=0,
xanchor="left",
yanchor="top",
pad=dict(t=50),
currentvalue=dict(
visible=True,
# prefix="x = ",
xanchor="right"
)
)
]
)
fig.show()

